“We categorized the tumors based on their gene expression profiles. In one group, the therapy was notably more effective than in the other,” explains Marjolein Lansbergen. She is a PhD student affiliated with the Department of Medical Oncology and the Center of Experimental and Molecular Medicine at Amsterdam UMC and Cancer Center Amsterdam. Her research focuses on metastatic pancreatic cancer under the supervision of medical oncologist Hanneke van Laarhoven and molecular biologist Maarten Bijlsma. The therapy in question is FOLFIRINOX, a widely used combination chemotherapy for this type of cancer. “FOLFIRINOX is generally the best available therapy, but not everyone benefits from it,” Lansbergen notes. For treatment selection, it is crucial to accurately predict which patients will respond positively to the treatment or not.

A tumor's sensitivity to chemotherapy depends not only on DNA-level mutations; gene expression at the mRNA level plays a significant role. This expression dictates which genes are active and how the cell functions and responds to its environment—including its response to therapy. Different cell types and tumors have unique gene expression profiles. Comparing these profiles allows researchers to classify tumors into (RNA) subgroups. Lansbergen aimed to devisea classification system that best predicts a patients chances to respond to chemotherapy. This would enable doctors to tailor treatment decisions by distinguishing different patient groups based on their gene expression profiles.
For their study, Lansbergen and her colleagues collected tissue samples from patients prior to FOLFIRINOX treatment. This was done as part of the CPCT-02 study, a large-scale data collection initiative. Tumor size was monitored using CT scans, and survival data was recorded. The tissue samples were sent to Hartwig Medical Foundation for Whole Transcriptome Sequencing. Using the resulting mRNA data, Lansbergen tested several commonly used RNA subgroup classifiers.
“Ultimately, the classification between so-called classical and basal-like tumors aligned best with our data,” Lansbergen explains. Basal-like tumors are generally more aggressive than classical tumors and resemble basal-like tumors found in breast and bladder cancers.
“We observed clear differences in survival and tumor reduction between the two groups,” says Lansbergen. Among patients treated with FOLFIRINOX, those with classical tumors lived longer than those with basal-like tumors. Classical tumors also showed a higher likelihood of ceasing growth compared to basal-like tumors. “It’s gratifying that we were able to demonstrate this,” Lansbergen says enthusiastically.
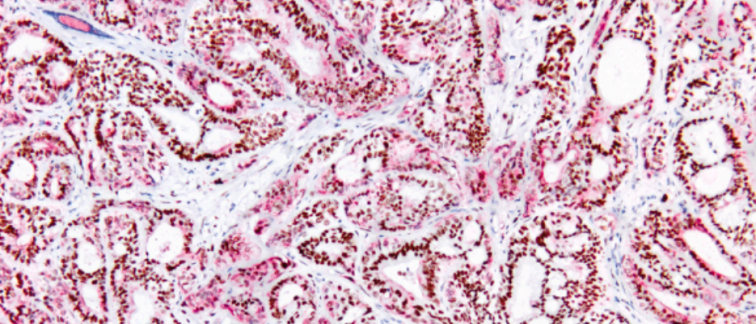
One important gene that exhibited differential expression was GATA6. This gene can be measured not only at the RNA level but also at the protein level using immunohistochemical GATA6 staining. Lansbergen elaborates: “GATA6 is a well-known marker associated with the classical tumor group. We observed that patients with high levels of GATA6 protein indeed lived longer on FOLFIRINOX compared to those with low levels of GATA6.”
In metastatic pancreatic cancer, a relatively simple protein staining can therefore be used to indicate the RNA subgroup of a tumor, helping to predict the effectiveness of treatment. Lansbergen concludes: “With this study, we demonstrate that gene expression profiles have the potential to support patients and doctors in making informed treatment decisions. Hopefully, this contributes to longer survival and fewer side effects.”
This research was recently published open access in Translational Research.
Hartwig Medical Foundation has built a database that includes RNAseq data from over 3,000 patients, encompassing the data used in the above study. An overview of all available data can be found on Hartwig’s website, where selections can be made based on various specifications such as primary tumor type, availability of RNA data and treatment type. An example display of this data overview is shown below. This tool was developed with support from the Dutch Cancer Society (KWF).